please consider which updates from the recently assigned tasks (both XG and PL) would be appropriate for you to briefly present at the meeting.
The meeting is tentatively scheduled for 10 am Fri.
This time is now confirmed.
RC (8/7): PMC-AT manuscripts page should be edited to indicate paper 1 in this case.
XG(8/8): Only Organizer can change the page name.
RC (8/7): Over the next month or so (until the continuous assay is finished) you should spend 2/3 of your time as priority finishing the continuous assay and
1/3 of time on the 2nd paper outlining.
Details:
a) given a 2/3 time commitment, please provide/confirm the estimated time for continuous assay completion including validation of the method (e.g. by comparison to FdL results).
XG(8/7):
Large Scale purification of protein PNCA, protein characterization, and activity measurement -----3 weeks.
Validation of the continuous assay----- 2-3 weeks.
b) 2nd paper: points below should start being incorporated as bullet points into appropriate sections of paper draft provided. If any questions, ask. This includes points that map rate constants previously
determined experimentally to our diagram, differences between base exchange rate constant determination method of Sauve and ours - assumptions he made, etc. Bibliography and citations should be started.
This will give me a better idea of length. A major focus of this paper is presentation of the model for sirtuin reaction kinetics, which can be used for either mechanism-based inhibitor and activator design and which was not presented in 1st paper. We will assume PCB as journal for now. Based on the length estimate, we will decide whether to move the derepression activation sections to a separate paper.
c) 2nd paper: some relevant sentences from 1st paper intro (including references, e.g. to NAD+ regulation of sirtuins) should be added where indicated in outline
d) 2nd paper: should add point to intro regarding how allosteric activators work only with SIRT1 and also only with certain substrates with hydrophobic +1/+6 residues. Provide some info on how many SIRT1 substrates do not have this feature, as motivation for other approaches.
e) 2nd paper: we should indicate early on that the paper deals with both mechanism-based inhibitor and activator design. PNAS paper on Ex-527 should be referenced here.
f) Lower priority: after some of above finished, we will discuss what is required to run the cell bio assays for upregulation and downregulation of sirtuin activity in our lab.
g) For relative inhibition section, more detail on the relative inhibition assay is needed. See below. Then add the latest data and protocol to the appropriate section. Some changes to our assays may be needed in the future to account for generation of NAM during reaction.
Prior to further studies on dihydropyridines, we will calculate the [NAM] over time in the endpoint activity assays. Will let you know when.
h) RC will review the list of leads identified through similarity search, especially the substructures used, prior to experiments with those leads. Assuming confirmation, experiments will start with inhibitor leads. These results may be presented in a third paper.
-Please provide lead time for delivery of the molecules identified through similarity search.
XG(8/7): It depends on the company we order from. In general, the chemicals will be shipped out within 10 business days after the order is confirmed.
-Please indicate which substructures were used to generate each lead.
-Please provide an estimate of time required for IC50 determinations for each lead using continuous assay. Hits will be passed to kinetic characterization (initial deacetylation rate) experiments.
XG(8/7):
Solubility test (in H2O or DMSO in common or specific solvent)---3 lead/day;
Optimization of range of the [inhibitor]---need 2-3 runs (low, medium, and high [ ]);
Background signal adjustment if compound has color---3 lead/day.
A 96-well plate can be run on TECAN or Fluoroskan for up to 4 compounds ---2 plates/day.
Data analysis will be performed from every run to optimize the final IC50---2 runs/day.
i) Please verify whether any complexes may be amenable to ITC binding affinity measurement (follow up on your group mtg task on this).
XG(8/7): We discussed the possibility of using ITC to bring in solid binding affinity data before. If the modulator potent enough, ITC can be applied (MST was used in PNAS paper for Ex527:NAD/substrate/ Apo-enzyme).
For accurate measurement, the comfortable zone will be 1nM - 100uM.
Time: One run for couple of hours. Then system cool down. For each binding pair, if everything goes smoothly, one would need at least two samples (1) with small molecule, (2) without small molecule_ reference. The concentrations of protein and/or small molecules need to be adjusted for best S/N, which would take more time. Typically people normally plan for 2- day experiment for each binding pair.
j) regarding base exchange assay, we need info on: price of custom-labeled reagents (e.g. NAM), whether reagent vendors can ship to us without any validation of our ability to handle such reagents, price of LSC, whether LSC can be purchased by any lab, confirmation that no local approval is needed for us to use such reagents.
k) 2nd paper: In PMC AT Research folder, there is a Hubbard 2014 review on sirtuin modulators that discusses inhibitor applications. Please add notes from Hubbard review on SIRT1 modulators (2014) regarding applications of inhibitors as bullet pts to paper draft. There is also a review from 2011 there.
l) Please look up any/all patents on dihydropyridines as drugs - there appear to be many used as calcium channel blockers.
Also please verify that no other other work has been done with these on sirtuins.
RC (8/6): Before proceeding to answer my new questions below posted today, please indicate approximately how long it will take you to complete each of the tasks, so we may develop a schedule. XG(8/1/14): Remaining tasks/questions
(1) you mentioned that ”Previous studies have focused on the complete steady-state kinetic mechanism (a) the inherent differences in catalytic efficiency and substrate preference among Sir2 enzymes (ySir2, yHST2, and SIRT2) for various histone peptides; (b) the overall kinetic mechanism; (c) Resolve the individual chemical steps of the Sir2 reaction.”specifically in terms of the rate constants of the reaction mechanism, what does mean? In what way were each of the individual chemical steps resolved? Subsequently, you mention that rapid quench was required to determine particular reaction rates.
XG(8/4): Previous studies have focused on resolving the steady-state kinetic mechanism. In terms of the rate constants, some of the individual step(s) have been measured experimentally or computationally. (A) Substrate Binding: Using equilibrium dialysis ([3H]AcH3), the binding of NAD+ to HST2 and the binding of acetylated substrate to HST2 were measured. The results supported that the acetylated peptide is first to bind with a Kd of 150 uM. No NAD+ binding was detected. Borra MT et al, Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases.(2004) Biochemistry 43: 9877-9887. (B) Transition State: A novel intermediate-ADPR-peptidyl-imidate intermediate-alpha-1’-O-alkylamidate intermediate has been identified. Asynchronous SN2 mechanism of NAD+ with acetyllysine nucleophile was discussed. X-ray structure data of the transition type analogues on the active site of sirtuins and a QM/MM study of Sir2Tm support this concept. Hawse, WF et al. Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure (2008) 16: 1368-1377. Hu, P et al. Highly dissociative and concerted mechanism for the nicotinamide cleavage reaction in Sir2Tm enzyme suggested by ab initio QM/MM molecular dynamicssimulations. J Am Chem Soc 2008, 130:16721-16728. A computational study of the Sir2Af2 catalyzed reaction of NAD+ with acetyllysine reached a similar conclusion, fortified with experimental kinetic isotope effects (KIE) data (calculated for candidate transition state structure using computational methods like Gaussian 03 and ISOEFF 98). Cen, Y et al. Transition state of ADP-ribosylation of acetyllysine catalyzed by Sir2AF2 determined by KIE and computational approaches. JACS(2010) 132:12286-12298. (C) Base Exchange: Using HPLC with [Carbonyl-14C]NAM and 18O-NAM, the base exchange reactions on ySir2, Sir2Af, and mSir2 (SIRT1) were studied. A strategy for increasing Sir2 enzyme catalytic activity in vivoby inhibition of chemical exchange. NAM and isoNAM activation of Sir2 deacetylase activity is achieved without affecting substrate or NAD+ binding by altering the proportion of imidate-enzyme complexes proceeding toward the deacetylated product. A.A. Sauve, V.L. Schramm, Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry, Biochemistry 42 (2003)9249–9256. A.A. Sauve, R.D. Moir, V.L. Schramm, I.M. Willis, Chemical activation of Sir2- dependent silencing by relief of nicotinamide inhibition, Mol. Cell 17 (2005) 595–601. (D) Spontaneous non-enzymatic equilibration 2’AADPR_3’AADPR:The mechanism of acetyl transfer to NAD+ includes (1) ADP ribosylation of the peptide acyl oxygen to form a high-energy O-alkyl amidate intermediate, (2) attack of the 2’-OH group on the amidate to form a 1’,2’-acyloxonium species, (3) hydrolysis to 2’-AADPR by the attack of water on the carbonyl carbon, and (4) an SIR2-independent transesterification equilibrating the 2’- and 3’-AADPRs. Approachs used:1H NMR, 2D NMR, MS/MS, 18O exchange, and HPLC. Sauve AA et al. Chemistry of Gene Silencing: The Mechanism of NAD+-Dependent Deacetylation Reactions. Biochemistry (2001) 40: 15456-15463. Smith, BC et al. Sir2 protein deacetylases: Evidence for chemical intermediates and functions of a conserved histidine. Biochemistry (2006) 45: 272-282.__ RC: We will need to indicate which rate constants in the diagram these correspond to. In general, please try to map what you are reading to the models we are developing (not enough to only quote the relevant literature).
XG(8/6): map_reported sirtuin kinetic studies.pptx
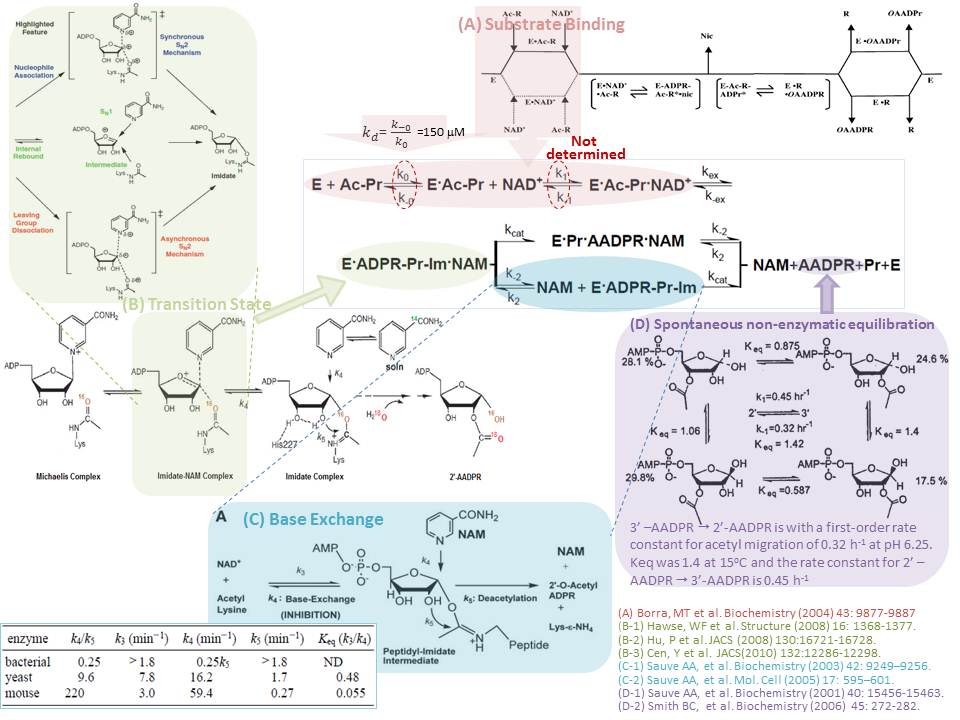
RC: Please indicate mapping of kex/k-ex
Note there is a section of paper indicating different methods of obtaining rate constants (including QM/MM for kex). The relevant papers should be cited there with notes.
References can be found in dropbox/PMC AT Research/Guan/08.04.14 wiki post
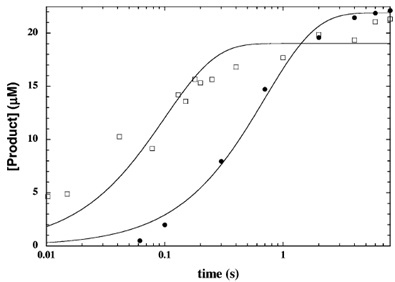
(2) you mentioned “To obtain the rate constant (k), the plot of product concentration formed over time was fitted to a single-exponential equation and then After formation of the ternary complex, nicotinamide is cleaved at a rate of 7.3 s-1 and subsequent formation of OAADPr occurs at 1.3 s-1. “ What is the relationship between these two rate constants and the rate constant k mentioned above?
XG(8/4): The NAM and OAADPr formation rate constants are 7.3 and 1.3 s-1 respectively. They are calculated from the plot using aforementioned method.RC: Not sure we are talking about the same thing here. There was a rate constant k you mentioned. Now you mentioned two rate constants. Do you mean that a single exponential fit is done for the formation of two different products and then two different k's are obtained? Please indicate in the diagram which rate constants we are referring to in each case.
XG(8/6): Two exponential fits are done for the formation of NAM and OAADPR respectively. Two different k’s are obtained. Using a quench-flow apparatus, the HST2 reaction under single-turnover conditions was monitored.
- To determine k_NAM (open square), [14C]NAD+ was used. Fractions from reversed-phase HPLC were subjected to scintillation counting to determine the amount of [14C]nicotinamide formed and [14C]NAD+ left. An average rate of 7.3 s-1 was determined from three separate experiments.
- To determine k_OAADPR (solid circle), [3H]AcH3 was used as a substrate. The amount of OAADPR formed and the amount of nicotinamide formed vs log time were fitted into a single-exponential equation to determine the rate of product formation. An average rate of 1.26 s-1 was yield from two experiments.
(3) Regarding HPLC, you mentioned at the group meeting that this could be used in post-steady state kinetics (i.e., the transient kinetics that occur after the initial stages of the reaction where steady state conditions no longer hold). Can you provide more details? These discussions will eventually need to be integrated with the relevant kinetic characterization sections of the outline I sent for paper 2.
XG(8/4): HPLC-based Deacetylase Assay relies on the separation of substrates and products of the deacetylase reaction by a reversed-phase HPLC. Quenched reaction mixtures are injected onto a C18 column and, using a gradient of increased levels of organic solvent, substrates, products, and enzyme can be resolved. This assay is applicable to all three classes of deacetylases. For characterization of most Sir2-like enzymes, monoacetylated H3 and H4 peptides, corresponding to the 20 N-terminal residues of histone H3 and H4, can be used. The 110-ul reactions are carried out at 37oC in 50 mM Tris (or phosphate), pH 7.5, with 1 mM DTT. NAD+ and acetylated peptide are mixed and pre-incubated in a 37oC water bath for 5 min. Typical concentrations of NAD+ and acetylated peptide have ranged from 0.25 uM to 1 mM; however, the range should be determined empirically, as the Km values for these substrates may differ by orders of magnitude depending on the Sir2 homologue. The reaction is initiated by the addition of enzyme. The reaction is quenched by the addition of TFA to a final concentration of 1%. Quenched samples are kept on ice or stored in -20oC if not immediately injected onto the HPLC column. Samples are injected onto a reversed-phase HPLC column (e.g., a Vydac C18 column, 4.6 X 250 mm, 201SP104) to resolve substrates and products. A 100-ul loop is typically used for the injections and the flow rate is set to 1 ml/min. After injection, the system is run isocratically with solvent A (0.05% TFA/H2O) for 1 min followed by increasing levels of solvent B (0.02% TFA in acetonitrile). The gradient used for each assay may vary depending on the type of peptide substrate used. For efficient separation of reactions containing the 20-mer AcH3 N-terminal peptides, a gradient of 0–20% B over 20 min is used. For efficient resolution of reactions containing the 20-mer H4 peptides, the following gradient is used: 0–10% solvent B for 4 min followed by 10–25% B for 25 min. Following each run, the column is washed with 100% B for 3–5 column volumes followed by re-equilibration with solvent A for 3–5 column volumes. Elution of substrates and products is monitored by measuring the absorbance at 214 nm (to monitor all substrates and products) or at 260 nm (to specifically monitor nicotinamide, NAD+, and OAADPr). Using the above gradients, a good resolution between the monoacetylated and deacetylated peptides can be achieved. Deacetylated H3 peptide typically elutes at 16 min, acetylated H3 at 18 min, deacetylated H4 at 15 min, and acetylated H4 at 17 min. Substrates and products elute at the approximate percentages of solvent B: nicotinamide at 5%, deacetylated H3 at 16%, deacetylated H4 at 13%, acetylated H3 at 18%, and acetylated H4 at 14%, NADþ at 12%, and OAADPr at 8%. The areas of the peaks are integrated for quantification. To calculate the percent deacetylation, the area of the deacetylated peptide peak is compared to the combined areas of the acetylated and deacetylated peptide peaks. Because a known amount of acetylated peptide is used, the percentage of the deacetylation is then used to determine the amount of deacetylated product formed over the particular time of the assay, to obtain an initial rate.
This HPLC-based assay is also applicable for separating radiolabeled substrates and products. For example, [3H]acetylated histone peptides or [14C]- or [32P]-labeled NADþ can be utilized. [14C]NAD+ can be synthesized using the Sir2-catalyzed nicotinamide-NAD+ exchange procedure (described later) or purchased commerically, and [32P]NAD+ can be obtained from NEN Life Science Products (800 Ci/mmol). The amounts of the radiolabeled substrates and products can be monitored and quantified by collecting the fractions eluted from the HPLC, adding a constant volume of each fraction into scintillation vials and determining the radioactivity of each fraction by scintillation counting. To calculate the amounts of product formed, the radioactivity of the product can be divided by the total radioactivity of all the fractions collected, which corresponds to the radioactivity of the substrate prior to the reaction. The percent product is then multiplied by the concentration of radiolabeled substrate to obtain the concentration of product formed. Alternatively, the total radioactivity of the product can be divided by the specific activity (CPM/mol) of the radiolabeled substrate to obtain the moles of product formed. One of the limitations of this HPLC-based assay is its relatively timeconsuming nature, where each HPLC run can take approximately 1 h. Only a limited number of injections can be performed over a typical working day. Use of an auto-injector can greatly facilitate the analysis of a large number of samples. Another limitation of this assay is the detector’s inability to detect low concentrations of substrates when absorbance is used to quantitate levels of deacetylation. With low peptide concentrations (below 5 uM) larger injection volumes (e.g., 2 ml) are required in order to have sufficient substrate and products to accurately detect. Moreover, because the retention times may change over column useage, it is necessary to check the elution of standards routinely.
(4) Note that the approach to kinetic characterization in our 2nd paper outline provides a quantitative method for calculating all the rate constants represented in our reaction scheme (which omits certain chemistry and product dissociation steps). Given the posting above, can you consider how to insert bullet points regarding the literature methods in the appropriate sections of the outline that demonstrates how our method offers this advantage? E.g., Sauve's base exchange methods involve various approximations that we do not employ. At least one other slide shows the assumption Kd=Km, which we do not apply. XG(8/6): 1 day
(5) Please consider bullet points that can be added to the outline regarding the disease applications of inhibitors. XG(8/6): 1-2 days.
(6) Recall the question regarding the cost of HTS. I was referring to CRO HTS wherein there is a cost quoted per well. This depends on the application, but we can check some approximate prices. Are our assays capable of being run by a CRO in higher throughput, or are they too complicated?
XG(8/4): Few well established companies provide such services. Here two of such companies are listed, Charles River Libraries and Jackson Labs. Here are the services provided by Charles River Libraries:
- Target discovery & validation: Hit identification / Hit-to-lead/Lead optimization / Academic engagement initiative (Grant application support, track record)
- Assay development & screening: Biochemical to Phenotypic assay development/ cell line generation / HTS/High-content screening/ compound screening libraries/orthogonal screening platforms/ion Channel screening/fragment-based drug discovery/novel biomarker identification
- Synthetic & medicinal chemistry: Medicinal chemistry/computer-aided drug design/compound library design/synthetic chemistry/analytical chemistry/process & scale-up chemistry
- Structural biology & biophysics: protein production/protein crystallization &x-ray crystallography/crystal bank/biophysics/fragment-based drug discovery
- Pharmaceutical sciences & formulation: analytics & structural analysis, physical property measurement, solubility, dissolution & stability, salt screening, polymorphism screening, formulation & excipient compatibility.
(7) Regarding the literature on HTS for sirtuins, for the major classes of sirtuin inhibitors/activators currently in pharma development, please list how they were discovered (library type, size), approximate cost based on the prices above, and how they are being optimized (if any info is available, including what types of substitutions are being screened). (8) Please see the paper in PMC-AT Research dropbox called SRT2104_mouse_lifespan_2014. Can a CRO do preclinical studies of this type?
(9) Indicate when the continuous assay will be ready XG: - PCR amplification of PNCA from Samonella genomic DNA (Primer design, PCR condition optimization, …) - Subcloning into appropriate expression vector-pGEX6P3. (TOPO cloning, transformation of TOP10 chemical competent cell, identification of positive clones, Mini and Midiprep for sequencing, Digestion, Alkaline phosphatase, Calf Intestinal_CIP for Ligation, …) - Plasmid maxi preparation - Linearism the constructs - Transformation the linear construct into the appropriate host strain BL21(DE3) - Confirmation of positive transformants - Expression optimization and confirmation. (Screening of transformants, media formulation and inducer concentrations, induction temperature and length, culture lysate conditions, SDS-PAGE gel, …) - Large-scale expression. - Purification and confirmation. - Activity measurements RC: Time required? XG(8/4): The experiment was stopped at the purification step. It will take 2 weeks for purification and confirmation and 1 week for activity measurements.
(10)Indicate the approximate amount of time that would be required to get the base exchange assays set up XG: Most time consuming of the set up process is the purchasing of Liquid scintillation analyzer needed for the experiment. Other than that, we will need a separate room for storage and handling of radioactive reagents. Lab members need to be trained as well (can go on line and check where this kind of training is provided).
(11)Provide a pdf of the final paper with inline figures on the dropbox. RC: Please prepare a schedule for the above. 11 should be done early next week. XG(8/4): pdf file named as "PLOS ONE resubmission with figures and tables_08.04.2014" has been created in the dropbox. Dropbox/PMC-AT Manuscript/PCB-SIRT3 inhibition/PLOS One resubmission/